DETECT Model:

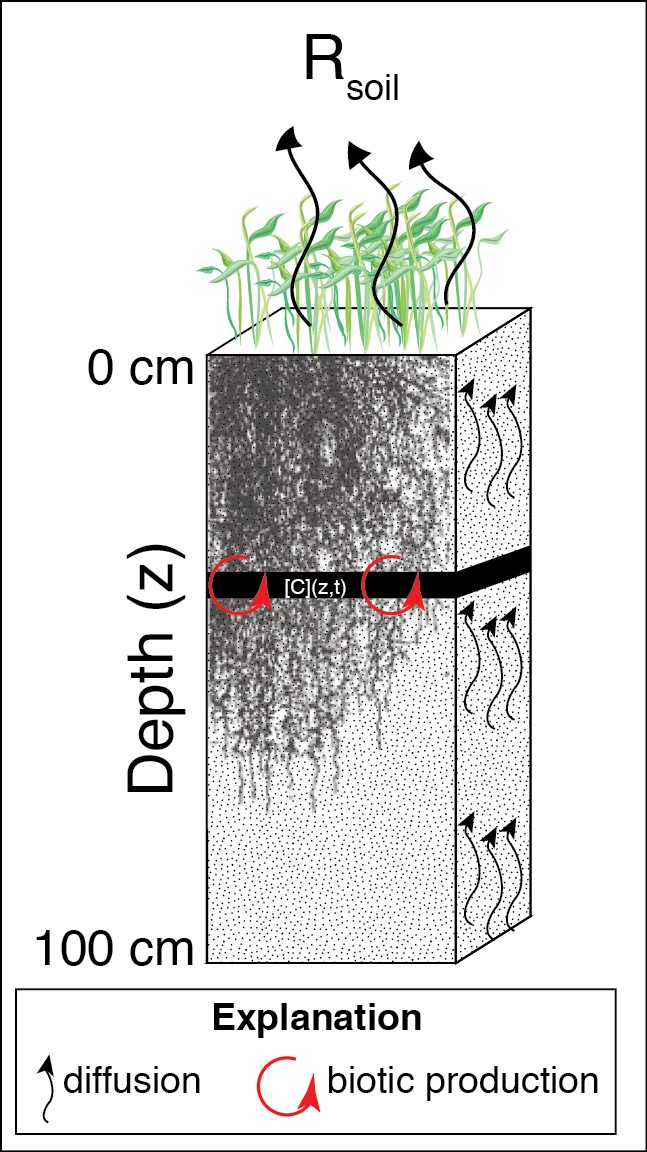 The DETECT model simulates CO2 production by roots and microbes at 100 0.01-m depth intervals at 6-hourly time increments, but it is flexible enough to accommodate finer or coarser depth and time intervals. The backbone of DETECT is a partial differential equation (PDE) model motivated by diffusion theory, and solved numerically. In Ryan et al. (2018) and Samuels-Crow et al. (2018), we used the DETECT model to simulate time- and depth-varying soil CO2 concentration and CO2 production rates and time-varying surface soil CO2 efflux (Rsoil) over a 183-day growing season in a mixed-grass prairie. The model is driven by environmental data collected at the Prairie Heating and CO2 Enrichment (PHACE) experiment near Cheyenne, Wyoming. PHACE data were also used to derive realistic parameter values associated with the submodels for CO2 production and diffusivity, and additional data were used to qualitatively evaluate model predictions.
The DETECT model simulates CO2 production by roots and microbes at 100 0.01-m depth intervals at 6-hourly time increments, but it is flexible enough to accommodate finer or coarser depth and time intervals. The backbone of DETECT is a partial differential equation (PDE) model motivated by diffusion theory, and solved numerically. In Ryan et al. (2018) and Samuels-Crow et al. (2018), we used the DETECT model to simulate time- and depth-varying soil CO2 concentration and CO2 production rates and time-varying surface soil CO2 efflux (Rsoil) over a 183-day growing season in a mixed-grass prairie. The model is driven by environmental data collected at the Prairie Heating and CO2 Enrichment (PHACE) experiment near Cheyenne, Wyoming. PHACE data were also used to derive realistic parameter values associated with the submodels for CO2 production and diffusivity, and additional data were used to qualitatively evaluate model predictions.
The PDE that underlies the DETECT model (v1.0) accounts for time- and depth-varying CO2 diffusivity and CO2 production by root and microbial respiration. We used a pair of PDEs, one describing the soil CO2derived from root respiration (subscripted with R), and the other for CO2 derived from microbial respiration (M) such that for K = R or M:

cK(z,t) is CO2 concentration (mg CO2 m-3), Dgs(z,t) is the effective diffusivity of CO2 through the soil (m2/s), and SK(z,t) is the source (or production) term (mg CO2 m-3) all of which vary by depth z(meters) and time t (hours). Total soil CO2 and total CO2 production are given as c(z,t) = cM(z,t) + cR(z,t) and S(z,t) = SM(z,t) + SR(z,t), respectively.
Model Download:
Instructions:
The Model Download file above includes the DETECT v1.0 manual along with all the scripts and input data necessary to run the model using parameter values and environmental driving data from the Wyoming PHACE site.
File Description:
ScriptFile.m — This file will run the DETECT model and save all outputs. It is currently set up to run the model based on the PHACE site parameters and driving data and must be modified based on the manual’s instructions to run it with user data. In order to run the PHACE site example, simply open and run this file.
SourceTerm.m — This file is a Matlab function that calculates microbial and root respiration (and total production).
DETECTmodel.m — This file is a Matlab function that calculated diffusivity and uses it, along with production calculated by the SourceTerm function, to calculate soil respiration.
Inputs\PHACE6hourly.mat — This file is the 6-hourly input data from the PHACE site. This file was made using the \Data_6hourly_Creator.m and the PHACE data (included as .csv files).
Outputs\ — This folder includes the output from running ScriptFile.m based on the PHACE data
Plots\ — Rsoil_timeseries.m and soilCO2_timeseries.m make time series plots of DETECT model output from the PHACE site along with measurements from the site as in figures 2 and 5 of Ryan et al. (2018).
JGR Example
Instructions:
The JGR Example directory includes driving data model output for each of the six simulation experiments described in Samuels-Crow et al. (2018). For convenience, we have included select driving data (e.g., soil moisture and temperature) in both the driving data and output files. The ReadMe file (included in the directory) provides further details about the contents of each file along with instructions for running DETECT using the driving data files.
References:
- Ryan, E., Ogle, K., Kropp, H., Samuels-Crow, K., Carrillo, Y., and Pendall, E. (2018). Modelling soil CO2 production and transport with dynamic source and diffusion terms: Testing the steady-state assumption using DETECT v1.0. Geoscientific Model Development. https://doi.org/10.5194/gmd-2017-223 abstract
- Samuels-Crow, K., Ryan, E., Pendall, E., Ogle, K. (2018). Temporal coupling of subsurface and surface soil CO2 fluxes: Insights from a non-steady state model and cross-wavelet coherence analysis, Journal of Geosphysical Research – Biogeosciences. DOI:10.1002/2017JG004207abstract
